Can we use the modulation of the immunosensors OAS and cGAS for the development of new drugs against infectious and inflammatory diseases?
What is this research project about?
The innate immune sensors activate interferon-driven antiviral responses upon recognition of pathogen-associated molecular patterns (PAMPs) and serve as a rheostat for the metabolic activity of the microbiota and its exposure to diet, xenobiotics, and infections. The ability to modulate innate immune sensors opens new ways to novel anti-viral and anti-inflammatory drugs, and therapies against cancer and many aging-associated metabolic, neoplastic, autoimmune or autoinflammatory disorders. The cGAS/OAS family of innate immune sensors is among the most promising targets for the development of new antimicrobial and immunomodulatory agents. cGAS/OAS share a highly conserved structure of the active sites with other nucleotide triphosphate transferases (NTPTs). Targeting active sites of cGAS/OAS may interfere with many important biological processes through unspecific inhibition (cross-reactivity). Allosteric regulation can solve the cross-reactivity problem and help develop specific activity modulators of the innate immune sensors.
What’s the current status?
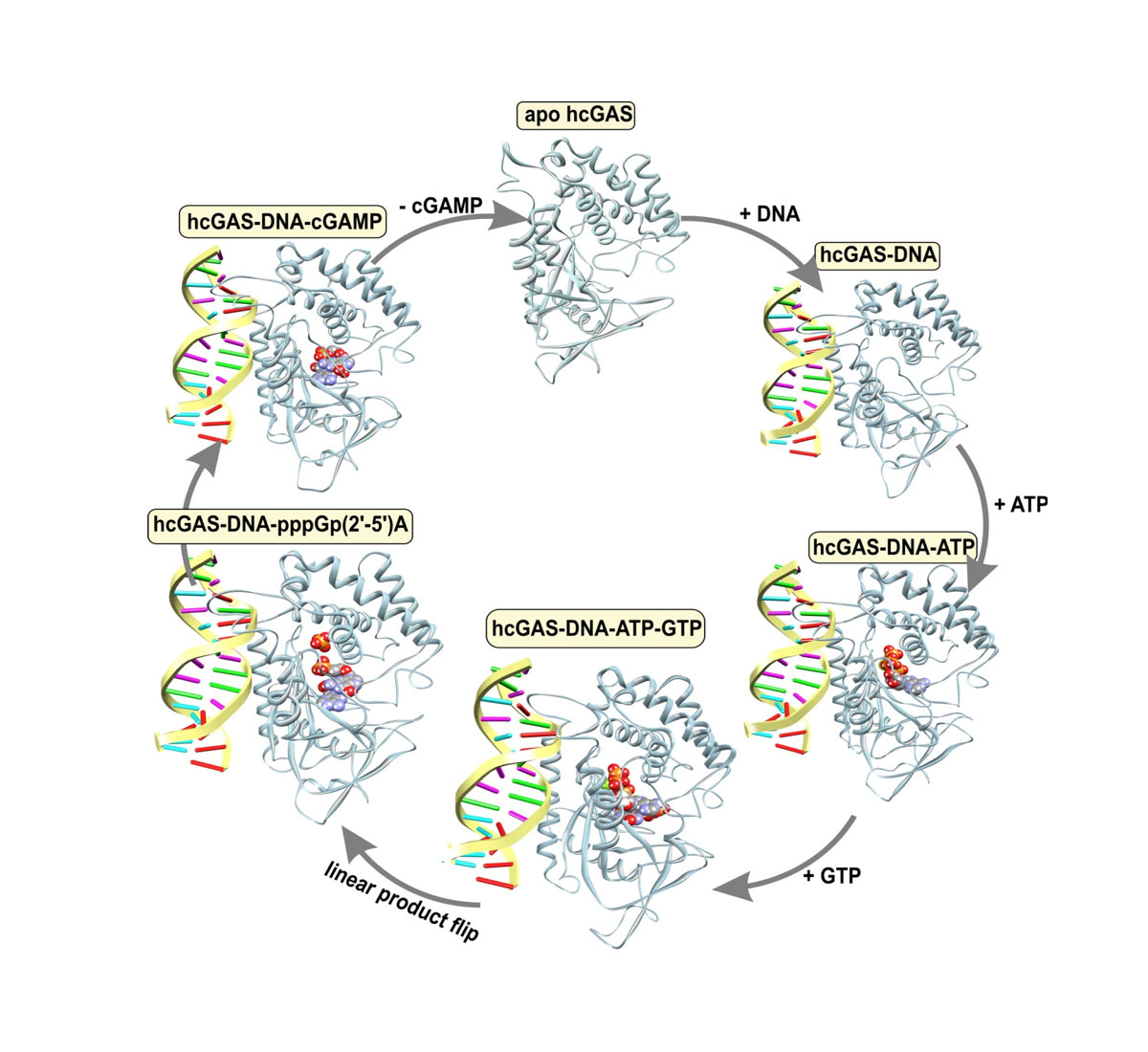
Recently, we established the detailed mechanisms of nucleic acid-induced activation of OAS1 and cGAS, the individual roles of nucleic acids and substrates, the sources of 2’-specificity of product formation, and the basis of functional divergence between OAS1 and cGAS. We also developed a new method that allows the rational identification of allosteric sites for the specific regulation of enzymes with highly-conserved catalytic centers, and successfully demonstrated its applications. These data and methodology are currently being used to discover and test the potency of allosteric pathways as targets for modulating the activity of cGAS/OAS.
How do we get there?
To reveal communication pathways that connect the catalytic centers of innate immune sensors with suitably positioned allosteric pockets, we are dissecting the complete enzymatic cycles of cGAS and OAS1, using approaches that were successfully applied in our previous works on UDP-sugar pyrophosphorylases1,2 and myosin isoforms3,5. The available structural information will be further refined to obtain more detailed information about cGAS dynamics and allostery. Allosteric sites and communication pathways will be tested and validated by the generation of mutant constructs that bias the positioning of active structural elements in a well-defined manner. HPLC/MS-based assays have been established to measure the catalytic activity of wild-type and mutant constructs. Allosteric sites that pass the validation tests are used as targets for the development of small-molecule inhibitors or activators.