How can diagnosis be improved to achieve early identification of as many affected individuals as possible?
What is this research project about?
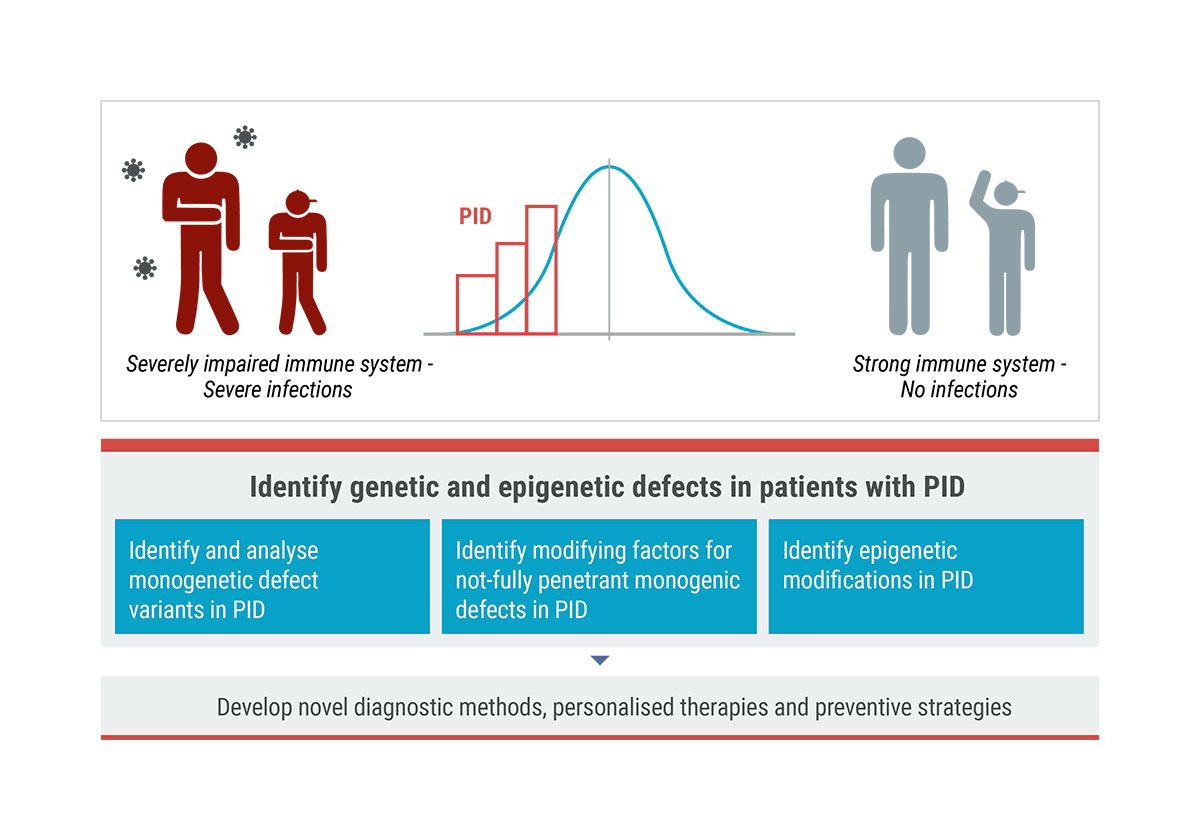
The fitness of the immune system is unevenly distributed in the population. On the one hand there are anecdotal reports on the “90-yr-old smoker, who has never been admitted to hospital”; on the other hand there are immunocompromised individuals who would not survive without medical intervention, sometimes necessitating even bone marrow transplantation. These individuals, with a profoundly impaired immune system, manifest with severe and often untypical (e.g. opportunistic) life-threatening infections and a predisposition to (mainly virally-induced) malignancies. In between these two extremes, however, there is a distribution of immune fitness, where patients with recurrent non-life threatening -but repeated or prolonged- infections, represent the majority of cases. In the last decades more than 350 different genes have been found to be mutated in immunodeficiency patients.
What’s the current status?
Depending on the cohort of study, the more than 350 genes associated with primary immunodeficiencies (PID), can only explain about 15-60% of cases. Certainly, there are probably more genes yet to be discovered, however, it is also likely that our search is not deep enough and that the monogenic model is insufficient to fulfill the diagnostic odyssey. Nowadays, Next-generation sequencing (NGS) technologies are routinely used in the clinic to obtain a genetic diagnosis. Most PID patients undergo targeted gene panel (TGP) sequencing or whole exome sequencing (WES), but still, a substantial number of them remain undiagnosed, as it is true for most Mendelian diseases. There may be many reasons for this; first, standard NGS bioinformatics pipelines are not specifically designed to detect copy number alterations (CNAs) or structural variants (SV), and these may be missed. Second, the use of TGP or WES limits the search to the coding part of the genome (2%). Third, other causative or contributing factors, which are excluded with the monogenic model, such as epigenetic profiles and the regulation of gene expression or the gut microbiome, have not been sufficiently studied.

More patients with immune defects could be registered.
What are the project goals?
The main objectives are:
- Reducing the number of unsolved cases, since the majority of patients with immunodeficiency are currently undiagnosed
- Improving our understating on how the epigenome works and its role in primary immunodeficiency disease.
- Understanding the relationship between the gut microbiome and disease severity and comorbidity in PID patients with malignancies, autoimmunity or inflammation
How do we get there?
The percentage of positively diagnosed patients could increase, first, by reanalyzing WES data with custom bioinformatics pipelines or with the use of novel alternative methods like next generation mapping (NGM) to detect CNAs or SVs, including insertions, translocations and inversions, which are not detectable by commonly used techniques. Second, by investigating epigenetic patterns of DNA methylation and histone modification in a genome-wide approach. Third, by sequencing the gut microbiome we can study the phylogenic and functional diversity of the gut microbiota and its role in shaping the immune system. Finally, microbiome diversity and co-morbidity in patients with PID could be addressed with the analysis of intestinal immunoglobulins.
Project A2 obtains and registers its patients from the national registry for immunodeficient patients, the PIDnet registry, which has existed since 2009. The PIDnet Registry is part of the European immunodeficiency registry of ESID (European Society for Immunodeficiencies). Since the PIDnet Registry is responsible for the clinical data of patients, the PIDnet Registry is an integral part of RESIST.In addition, the project includes data and biomaterials from the cohort of the Clinical Research Group 250 (KFO 250) in its research.

High-molecular DNA analysis to identify structural variants of the genome.
Projectleaders
Project title: Infection predisposition in primary immundeficiencies


Project A2 Publications
Publications 2024
Abnormal biomarkers predict complex FAS or FADD defects missed by exome sequencing. Rensing-Ehl A, Lorenz MR, Führer M, Willenbacher W, Willenbacher E, Sopper S, Abinun M, Maccari ME, König C, Haegele P, Fuchs S, Castro C, Kury P, Pelle O, Klemann C, Heeg M, Thalhammer J, Wegehaupt O, Fischer M, Goldacker S, Schulte B, Biskup S, Chatelain P, Schuster V, Warnatz K, Grimbacher B, Meinhardt A, Holzinger D, Oommen PT, Hinze T, Hebart H, Seeger K, Lehmberg K, Leahy TR, Claviez A, Vieth S, Schilling FH, Fuchs I, Groß M, Rieux-Laucat F, Magerus A, Speckmann C, Schwarz K, Ehl S; ALPS Study Group. J Allergy Clin Immunol. 2024 Jan;153(1):297-308.e12. doi: 10.1016/j.jaci.2023.11.006. Epub 2023 Nov 17. PMID: 37979702.
Epigenetic immune cell quantification for diagnostic evaluation and monitoring of patients with inborn errors of immunity and secondary immune deficiencies. Ramirez NJ, Schulze JJ, Walter S, Werner J, Mrovecova P, Olek S, Sachsenmaier C, Grimbacher B, Salzer U. Clin Immunol. 2024 Mar;260:109920. doi: 10.1016/j.clim.2024.109920. Epub 2024 Feb 1. PMID: 38307474.
Engineering Material Properties of Transcription Factor Condensates to Control Gene Expression in Mammalian Cells and Mice. Fischer, A. A. M., H. B. Robertson, D. Kong, M. M. Grimm, J. Grether, J. Groth, C. Baltes, M. Fliegauf, F. Lautenschläger, B. Grimbacher, H. Ye, V. Helms, and W. Weber. 2024. Small: e2311834.
Phenotypic and pathomechanistic overlap between tapasin and TAP deficiencies. Elsayed, A., S. von Hardenberg, F. Atschekzei, T. Graalmann, C. Jänke, T. Witte, F. C. Ringshausen, and G. Sogkas. 2024. J Allergy Clin Immunol.
Current genetic diagnostics in inborn errors of immunity. Sake, S. M., X. Zhang, M. K. Rajak, M. Urbanek-Quaing, A. Carpentier, A. P. Gunesch, C. Grethe, A. Matthaei, J. Ruckert, M. Galloux, T. Larcher, R. Le Goffic, F. Hontonnou, A. K. Chatterjee, K., Hardenberg, S., I. Klefenz, D. Steinemann, N. Di Donato, U. Baumann, B. Auber, and C. Klemann. 2024. Front Pediatr 12: 1279112.
B-cells absence in patients diagnosed as inborn errors of immunity: a registry-based study. Khoshnevisan, R., S. Hassanzadeh, C. Klein, M. Rohlfs, B. Grimbacher, N. Molavi, A. Zamanifar, A. Khoshnevisan, M. Jafari, B. Bagherpour, M. Behnam, S. Najafi, and R. Sherkat. 2024. Immunogenetics 76: 189-202.
PLCG2-associated immune dysregulation (PLAID) comprises broad and distinct clinical presentations related to functional classes of genetic variants. Baysac K, Sun G, Nakano H, Schmitz EG, Cruz AC, Fisher C, Bailey AC; PLCG2-Immune Dysregulation Working Group; Mace E, Milner JD, Ombrello MJ. J Allergy Clin Immunol. 2024 Jan;153(1):230-242. doi: 10.1016/j.jaci.2023.08.036. Epub 2023 Sep 26. PMID: 37769878
Publications 2023
Future Directions in the Diagnosis and Treatment of APDS and IEI: a Survey of German IEI Centers. Vanselow S, Hanitsch L, Hauck F, Körholz J, Maccari ME, Meinhardt A, Sogkas G, Schuetz C, Grimbacher B. Front Immunol. 2023 Oct 5;14:1279652.
A Toolkit for Monitoring Immunoglobulin G Levels from Dried Blood Spots of Patients with Primary Immunodeficiencies. Haberstroh H, Hirsch A, Goldacker S, Zessack N, Warnatz K, Grimbacher B, Salzer U. J Clin Immunol. 2023 Aug;43(6):1185-1192. doi: 10.1007/s10875-023-01464-0. Epub 2023 Mar 21.
JAKs and STATs from a Clinical Perspective: Loss-of-Function Mutations, Gain-of-Function Mutations, and Their Multidimensional Consequences Ott N, Faletti L, Heeg M, Andreani V, Grimbacher B. . J Clin Immunol. 2023 Aug;43(6):1326-1359.
Fecal Immunoglobulin Levels as a Modifier of the Gut Microbiome in Patients with Common Variable Immunodeficiency. Nöltner C, Bulashevska A, Hübscher K, Haberstroh H, Grimbacher B, Proietti M. J Clin Immunol. 2023 Aug;43(6):1208-1220.
Sequencing the B Cell Receptor Repertoires of Antibody-Deficient Individuals With and Without Infection Susceptibility.Lim YW, Ramirez NJ, Asensio MA, Chiang Y, Müller G, Mrovecova P, Mitsuiki N, Krausz M, Camacho-Ordonez N, Warnatz K, Adler AS, Grimbacher B. J Clin Immunol. 2023 Feb 24.
The link between rheumatic disorders and inborn errors of immunity. Sogkas G, Witte T. EBioMedicine. 2023 Mar 2;90:104501.
The GAIN Registry – a New Prospective Study for Patients with Multi-organ Autoimmunity and Autoinflammation. Staus P, Rusch S, El-Helou S, Müller G, Krausz M, Geisen U, Caballero-Oteyza A, Krüger R, Bakhtiar S, Lee-Kirsch MA, Fasshauer M, Baumann U, Hoyer BF, Farela Neves J, Borte M, Carrabba M, Hauck F, Ehl S, Bader P, von Bernuth H, Atschekzei F, Seppänen MRJ, Warnatz K, Nieters A, Kindle G, Grimbacher B. J Clin Immunol. 2023 Apr 21:1–13.
Activated Phosphoinositide 3-Kinase δ Syndrome: Update from the ESID Registry and comparison with other autoimmune-lymphoproliferative inborn errors of immunity. Maccari ME, Wolkewitz M, Schwab C, Lorenzini T, Leiding JW, Aladjdi N, Abolhassani H, Abou-Chahla W, Aiuti A, Azarnoush S, Baris S, Barlogis V, Barzaghi F, Baumann U, Bloomfield M, Bohynikova N, Bodet D, Boutboul D, Bucciol G, Buckland MS, Burns SO, Cancrini C, Cathébras P, Cavazzana M, Cheminant M, Chinello M, Ciznar P, Coulter TI, D’Aveni M, Ekwall O, Eric Z, Eren E, Fasth A, Frange P, Fournier B, Garcia-Prat M, Gardembas M, Geier C, Ghosh S, Goda V, Hammarstrom L, Hauck F, Heeg M, Heropolitanska-Pliszka E, Hilfanova A, Jolles S, Karakoc-Aydiner E, Kindle GR, Klemann C, Koletsi P, Koltan S, Kondratenko I, Körholz J, Krüger R, Jeziorski E, Levy R, Le Guenno G, Lefevre G, Lougaris V, Marzollo A, Mahlaoui N, Malphettes M, Meinhardt A, Merlin E, Meyts I, Milota T, Moreira F, Moshous D, Mukhina A, Neth O, Neubert J, Neven B, Nieters A, Nove-Josserand R, Oksenhendler E, Ozen A, Olbrich P, Perlat A, Pac M, Schmid JP, Pacillo L, Parra-Martinez A, Paschenko O, Pellier I, Sefer AP, Plebani A, Plantaz D, Prader S, Raffray L, Ritterbusch H, Riviere JG, Rivalta B, Rusch S, Sakovich I, Savic S, Scheible R, Schleinitz N, Schuetz C, Schulz A, Sediva A, Semeraro M, Sharapova SO, Shcherbina A, Slatter MA, Sogkas G, Soler-Palacin P, Speckmann C, Stephan JL, Suarez F, Tommasini A, Trück J, Uhlmann A, van Aerde KJ, van Montfrans J, von Bernuth H, Warnatz K, Williams T, Worth AJ, Ip W, Picard C, Catherinot E, Nademi Z, Grimbacher B, Forbes Satter LR, Kracker S, Chandra A, Condliffe AM, Ehl S; European Society for Immunodeficiencies Registry Working Party. J Allergy Clin Immunol. 2023 Jun 28:S0091-6749(23)00812-6.
ARPC5 deficiency leads to severe early-onset systemic inflammation and mortality. Sindram E, Caballero-Oteyza A, Kogata N, Chor Mei Huang S, Alizadeh Z, Gámez-Díaz L, Fazlollhi MR, Peng X, Grimbacher B, Way M, Proietti M. Dis Model Mech. 2023 Jul 1;16(7):dmm050145. doi: 10.1242/dmm.050145. Epub 2023 Jul 20. PMID: 37382373; PMCID: PMC10387347.
Clinical, immunological and molecular findings of 8 patients with typical and atypical severe combined immunodeficiency: identification of 7 novel mutations by whole exome sequencing. Alizadeh Z, Fazlollahi MR, Mazinani M, Badalzadeh M, Heydarlou H, Carapito R, Molitor A, de Oteyza ACG, Proietti M, Bavani MS, Shariat M, Fallahpour M, Movahedi M, Moradi L, Grimbacher B, Bahram S, Pourpak Z. Genes Immun. 2023 Aug;24(4):207-214. doi: 10.1038/s41435-023-00215-w. Epub 2023 Jul 29. PMID: 37516813.
Common Variable Immunodeficiency: More Pathways than Roads to Rome. Peng XP, Caballero-Oteyza A, Grimbacher B. Annu Rev Pathol. 2023 Jan 24;18:283-310. doi: 10.1146/annurev-pathmechdis-031521-024229. Epub 2022 Oct 20. PMID: 36266261.
Functional Relevance of CTLA4 Variants: an Upgraded Approach to Assess CTLA4-Dependent Transendocytosis by Flow Cytometry. Rojas-Restrepo J, Sindram E, Zenke S, Haberstroh H, Mitsuiki N, Gabrysch A, Huebscher K, Posadas-Cantera S, Krausz M, Kobbe R, Rohr JC, Grimbacher B, Gámez-Díaz L. J Clin Immunol. 2023 Nov;43(8):2076-2089. doi: 10.1007/s10875-023-01582-9. Epub 2023 Sep 23. Erratum in: J Clin Immunol. 2023 Oct 9;: PMID: 37740092; PMCID: PMC10661720.
GenIA, the Genetic Immunology Advisor database for inborn errors of immunity. Caballero-Oteyza A, Crisponi L, Peng XP, Yauy K, Volpi S, Giardino S, Freeman AF, Grimbacher B, Proietti M. J Allergy Clin Immunol. 2023 Nov 30:S0091-6749(23)01513-0. doi: 10.1016/j.jaci.2023.11.022. Epub ahead of print. PMID: 38040041.
Next generation sequencing (NGS)-based approach to diagnosing Algerian patients with suspected inborn errors of immunity (IEIs). Peng XP, Al-Ddafari MS, Caballero-Oteyza A, El Mezouar C, Mrovecova P, Dib SE, Massen Z, Smahi MC, Faiza A, Hassaïne RT, Lefranc G, Aribi M, Grimbacher B. Clin Immunol. 2023 Nov;256:109758. doi: 10.1016/j.clim.2023.109758. Epub 2023 Sep 9. PMID: 37678716.
Quality-of-life scores improve after 96 weeks of PEG-IFNa-2a treatment of hepatitis D: An analysis of the HIDIT-II trial. Dinkelborg K, Kahlhöfer J, Dörge P, Yurdaydin C, Hardtke S, Caruntu FA, Curescu MG, Yalcin K, Akarca US, Gürel S, Zeuzem S, Erhardt A, Lüth S, Papatheodoridis GV, Keskin O, Port K, Radu M, Celen MK, Idilman R, Weber K, Stift J, Wittkop U, Heidrich B, Mederacke I, von der Leyen H, Dienes HP, Cornberg M, Koch A, Manns MP, Wedemeyer H, Deterding K; HIDIT-2 Study Team. Liver Int. 2023 Aug;43(8):1663-1676. doi: 10.1111/liv.15602. Epub 2023 May 15. PMID: 37183524.
Telomere biology disorders may manifest as common variable immunodeficiency (CVID). Rolles B, Caballero-Oteyza A, Proietti M, Goldacker S, Warnatz K, Camacho-Ordonez N, Prader S, Schmid JP, Vieri M, Isfort S, Meyer R, Kirschner M, Brümmendorf TH, Beier F, Grimbacher B. Clin Immunol. 2023 Dec;257:109837. doi: 10.1016/j.clim.2023.109837. Epub 2023 Nov 8. PMID: 37944684.
ARPC5 deficiency leads to severe early-onset systemic inflammation and mortality. Sindram E, Caballero-Oteyza A, Kogata N, Chor Mei Huang S, Alizadeh Z, Gámez-Díaz L, Fazlollhi MR, Peng X, Grimbacher B, Way M, Proietti M. Dis Model Mech. 2023 Jul 1;16(7):dmm050145. doi: 10.1242/dmm.050145. Epub 2023 Jul 20. PMID: 37382373; PMCID: PMC10387347.
Publications 2022
Resolving the polygenic aetiology of a late onset combined immune deficiency caused by NFKB1 haploinsufficiency and modified by PIK3R1 and TNFRSF13B variants. Hargreaves CE, Dhalla F, Patel AM, de Oteyza ACG, Bateman E, Miller J, Anzilotti C, Ayers L, Grimbacher B, Patel SY. Clin Immunol. 2022 Jan;234:108910. doi: 10.1016/j.clim.2021.108910. Epub 2021 Dec 15. PMID: 34922003.
Phenotype, genotype, treatment, and survival outcomes in patients with X-linked inhibitor of apoptosis deficiency. Yang L, Booth C, Speckmann C, Seidel MG, Worth AJJ, Kindle G, Lankester AC, Grimbacher B; ESID Clinical and Registry Working Parties; Gennery AR, Seppanen MRJ, Morris EC, Burns SO. J Allergy Clin Immunol. 2022 Aug;150(2):456-466. doi: 10.1016/j.jaci.2021.10.037. Epub 2021 Dec 15. PMID: 34920033.
Multi-omics analysis of naïve B cells of patients harboring the C104R mutation in TACI. Ramirez N, Posadas-Cantera S, Langer N, de Oteyza ACG, Proietti M, Keller B, Zhao F, Gernedl V, Pecoraro M, Eibel H, Warnatz K, Ballestar E, Geiger R, Bossen C, Grimbacher B. Front Immunol. 2022 Aug 16;13:938240. doi: 10.3389/fimmu.2022.938240. PMID: 36072607; PMCID: PMC9443529.
Efficacy of dupilumab for the treatment of severe skin disease in cytotoxic T lymphocyte antigen-4 insufficiency: A role of type 2 inflammation? Arruda LK, Cordeiro DL, Langer SS, Koenigham-Santos M, Calado RT, Dias MM, Anhesini LR, Oliveira JB, Grimbacher B, Ferriani MPL. J Allergy Clin Immunol Glob. 2022 Sep 22;2(1):114-117. doi: 10.1016/j.jacig.2022.08.004. PMID: 37780100; PMCID: PMC10509893.
Detrimental NFKB1 missense variants affecting the Rel-homology domain of p105/p50. Fliegauf M, Kinnunen M, Posadas-Cantera S, Camacho-Ordonez N, Abolhassani H, Alsina L, Atschekzei F, Bogaert DJ, Burns SO, Church JA, Dückers G, Freeman AF, Hammarström L, Hanitsch LG, Kerre T, Kobbe R, Sharapova SO, Siepermann K, Speckmann C, Steiner S, Verma N, Walter JE, Westermann-Clark E, Goldacker S, Warnatz K, Varjosalo M, Grimbacher B. Front Immunol. 2022 Aug 29;13:965326. doi: 10.3389/fimmu.2022.965326. PMID: 36105815; PMCID: PMC9465457.
Confirmation of Hyperimmunoglobulin E Syndrome in Two Patients with an Ocular Problem: Detection of Two New DOCK8 Mutations. Saghafi S, Zandieh F, Fazlollahi MR, Glocker C, Frede N, Buchta M, Yang L, Mahmoudi AH, Houshmand M, Pourpak Z, Grimbacher B, Moin M. Iran J Allergy Asthma Immunol. 2022 Jun 18;21(3):355-363. doi: 10.18502/ijaai.v21i3.9809. PMID: 35822685.
Do common infections trigger disease-onset or -severity in CTLA-4 insufficiency? Krausz M, Mitsuiki N, Falcone V, Komp J, Posadas-Cantera S, Lorenz HM, Litzman J, Wolff D, Kanariou M, Heinkele A, Speckmann C, Häcker G, Hengel H, Gámez-Díaz L, Grimbacher B. Front Immunol. 2022 Nov 2;13:1011646.
Allele-Specific Disruption of a Common STAT3 Autosomal Dominant Allele Is Not Sufficient to Restore Downstream Signaling in Patient-Derived T Cells. König S, Fliegauf M, Rhiel M, Grimbacher B, Cornu TI, Cathomen T, Mussolino C. Genes (Basel). 2022 Oct 20;13(10):1912.
Common Variable Immunodeficiency-Associated Cancers: The Role of Clinical Phenotypes, Immunological and Genetic Factors. Bruns L, Panagiota V, von Hardenberg S, Schmidt G, Adriawan IR, Sogka E, Hirsch S, Ahrenstorf G, Witte T, Schmidt RE, Atschekzei F, Sogkas G. Front Immunol. 2022 Feb 17;13:742530.
Diagnostic Yield and Therapeutic Consequences of Targeted Next-Generation Sequencing in Sporadic Primary Immunodeficiency. Sogkas G, Dubrowinskaja N, Schütz K, Steinbrück L, Götting J, Schwerk N, Baumann U, Grimbacher B, Witte T, Schmidt RE, Atschekzei F. Int Arch Allergy Immunol. 2022;183(3):337-349.
Dysregulated PI3K Signaling in B Cells of CVID Patients. Harder I, Münchhalfen M, Andrieux G, Boerries M, Grimbacher B, Eibel H, Maccari ME, Ehl S, Wienands J, Jellusova J, Warnatz K, Keller B. Cells. 2022 Jan 28;11(3):464. doi: 10.3390/cells11030464.
Phenotypic spectrum in recessive STING-associated vasculopathy with onset in infancy: Four novel cases and analysis of previously reported cases. Wan R, Fänder J, Zakaraia I, Lee-Kirsch MA, Wolf C, Lucas N, Olfe LI, Hendrich C, Jonigk D, Holzinger D, Steindor M, Schmidt G, Davenport C, Klemann C, Schwerk N, Griese M, Schlegelberger B, Stehling F, Happle C, Auber B, Steinemann D, Wetzke M, von Hardenberg S. Front Immunol. 2022 Oct 6;13:1029423.
Copy Number Analysis in a Large Cohort Suggestive of Inborn Errors of Immunity Indicates a Wide Spectrum of Relevant Chromosomal Losses and Gains. Wan R, Schieck M, Caballero-Oteyza A, Hofmann W, Cochino AV, Shcherbina A, Sherkat R, Wache-Mainier C, Fernandez A, Sultan M, Illig T, Grimbacher B, Proietti M, Steinemann D. J Clin Immunol. 2022 Jul;42(5):1083-1092.
Publications 2021
High frequency of variants in genes associated with primary immunodeficiencies in patients with rheumatic diseases with secondary hypogammaglobulinaemia. Sogkas G, Dubrowinskaja N, Adriawan IR, Anim M, Witte T, Schmidt RE, Atschekzei F. Ann Rheum Dis. 2021 Mar;80(3):392-399.
A Pathogenic Missense Variant in NFKB1 Causes Common Variable Immunodeficiency Due to Detrimental Protein Damage. Fliegauf M, Krüger R, Steiner S, Hanitsch LG, Büchel S, Wahn V, von Bernuth H, Grimbacher B. Front Immunol. 2021 Apr 27;12:621503.
Bile acids regulate intestinal antigen presentation and reduce graft-versus-host disease without impairing the graft-versus-leukemia effect. Haematologica. Haring E, Uhl FM, Andrieux G, Proietti M, Bulashevska A, Sauer B, Braun LM, de Vega Gomez E, Esser PR, Martin SF, Pfeifer D, Follo M, Schmitt-Graeff A, Buescher J, Duyster J, Grimbacher B, Boerries M, Pearce EL, Zeiser R, Apostolova P. 2021 Aug 1;106(8):2131-2146. doi: 10.3324/haematol.2019.242990. PMID: 32675222; PMCID: PMC8327708.
Diagnostic Yield and Therapeutic Consequences of Targeted Next-Generation Sequencing in Sporadic Primary Immunodeficiency. Sogkas G, Dubrowinskaja N, Schütz K, Steinbrück L, Götting J, Schwerk N, Baumann U, Grimbacher B, Witte T, Schmidt RE, Atschekzei F. Int Arch Allergy Immunol. 2021 Oct 7:1-13. doi: 10.1159/000519199. Epub ahead of print. PMID: 34619682.
A novel NFKBIA variant substituting serine 36 of IκBα causes immunodeficiency with warts, bronchiectasis and juvenile rheumatoid arthritis in the absence of ectodermal dysplasia. Sogkas G, Adriawan IR, Ringshausen FC, Baumann U, Schröder C, Klemann C, von Hardenberg S, Schmidt G, Bernd A, Jablonka A, Ernst D, Schmidt RE, Atschekzei F. Clin Immunol. 2020 Jan;210:108269. 3,969 “doi: 10.1016/j.clim.2019.10826
Bowel Histology of CVID Patients Reveals Distinct Patterns of Mucosal Inflammation.van Schewick CM, Lowe DM, Burns SO, Workman S, Symes A, Guzman D, Moreira F, Watkins J, Clark I, Grimbacher B. J Clin Immunol. 2021 Oct 1. doi: 10.1007/s10875-021-01104-5. Epub ahead of print. PMID: 34599484.
Therapeutic options for CTLA-4 insufficiency. Egg D, Rump IC, Mitsuiki N, Rojas-Restrepo J, Maccari ME, Schwab C, Gabrysch A, Warnatz K, Goldacker S, Patiño V, Wolff D, Okada S, Hayakawa S, Shikama Y, Kanda K, Imai K, Sotomatsu M, Kuwashima M, Kamiya T, Morio T, Matsumoto K, Mori T, Yoshimoto Y, Dybedal I, Kanariou M, Kucuk ZY, Chapdelaine H, Petruzelkova L, Lorenz HM, Sullivan KE, Heimall J, Moutschen M, Litzman J, Recher M, Albert MH, Hauck F, Seneviratne S, Pachlopnik Schmid J, Kolios A, Unglik G, Klemann C, Snapper S, Giulino-Roth L, Svaton M, Platt CD, Hambleton S, Neth O, Gosse G, Reinsch S, Holzinger D, Kim YJ, Bakhtiar S, Atschekzei F, Schmidt R, Sogkas G, Chandrakasan S, Rae W, Derfalvi B, Marquart HV, Ozen A, Kiykim A, Karakoc-Aydiner E, Králíčková P, de Bree G, Kiritsi D, Seidel MG, Kobbe R, Dantzer J, Alsina L, Armangue T, Lougaris V, Agyeman P, Nyström S, Buchbinder D, Arkwright PD, Grimbacher B. J Allergy Clin Immunol. 2021 Jun 7:S0091-6749(21)00891-5. doi: 10.1016/j.jaci.2021.04.039. Online ahead of print. PMID: 34111452
There is no gene for CVID – novel monogenetic causes for primary antibody deficiency. Ramirez NJ, Posadas-Cantera S, Caballero-Oteyza A, Camacho-Ordonez N, Grimbacher B. Curr Opin Immunol. 2021 Jun 18;72:176-185. doi: 10.1016/j.coi.2021.05.010. Online ahead of print. PMID: 34153571 Review.
Pembrolizumab for treatment of progressive multifocal leukoencephalopathy in primary immunodeficiency and/or hematologic malignancy: a case series of five patients. Volk T, Warnatz K, Marks R, Urbach H, Schluh G, Strohmeier V, Rojas-Restrepo J, Grimbacher B, Rauer S. J Neurol. 2021 Jul 1. doi: 10.1007/s00415-021-10682-8. Online ahead of print. PMID: 34196768
TACI deficiency – a complex system out of balance. Salzer U, Grimbacher B. Curr Opin Immunol. 2021 Jul 8;71:81-88. doi: 10.1016/j.coi.2021.06.004. Online ahead of print. PMID: 34247095 Review.
Cellular and molecular mechanisms breaking immune tolerance in inborn errors of immunity. Sogkas G, Atschekzei F, Adriawan IR, Dubrowinskaja N, Witte T, Schmidt RE. Cell Mol Immunol. 2021 Apr 1:1-19. doi: 10.1038/s41423-020-00626-z. Online ahead of print. PMID: 33795850 Free PMC article. Review.
Beyond „Big Eaters“: The Versatile Role of Alveolar Macrophages in Health and Disease. Hetzel M, Ackermann M, Lachmann N. Int J Mol Sci. 2021 Mar 24;22(7):3308. doi: 10.3390/ijms22073308. PMID: 33804918 Free PMC article. Review.
A distinct CD38+CD45RA+ population of CD4+, CD8+, and double-negative T cells is controlled by FAS. Maccari ME, Fuchs S, Kury P, Andrieux G, Völkl S, Bengsch B, Lorenz MR, Heeg M, Rohr J, Jägle S, Castro CN, Groß M, Warthorst U, König C, Fuchs I, Speckmann C, Thalhammer J, Kapp FG, Seidel MG, Dückers G, Schönberger S, Schütz C, Führer M, Kobbe R, Holzinger D, Klemann C, Smisek P, Owens S, Horneff G, Kolb R, Naumann-Bartsch N, Miano M, Staniek J, Rizzi M, Kalina T, Schneider P, Erxleben A, Backofen R, Ekici A, Niemeyer CM, Warnatz K, Grimbacher B, Eibel H, Mackensen A, Frei AP, Schwarz K, Boerries M, Ehl S, Rensing-Ehl A. J Exp Med. 2021 Feb 1;218(2):e20192191. doi: 10.1084/jem.20192191. PMID: 33170215; PMCID: PMC7658692.
Biochemically deleterious human NFKB1 variants underlie an autosomal dominant form of common variable immunodeficiency. Li J, Lei WT, Zhang P, Rapaport F, Seeleuthner Y, Lyu B, Asano T, Rosain J, Hammadi B, Zhang Y, Pelham SJ, Spaan AN, Migaud M, Hum D, Bigio B, Chrabieh M, Béziat V, Bustamante J, Zhang SY, Jouanguy E, Boisson-Dupuis S, El Baghdadi J, Aimanianda V, Thoma K, Fliegauf M, Grimbacher B, Korganow AS, Saunders C, Rao VK, Uzel G, Freeman AF, Holland SM, Su HC, Cunningham-Rundles C, Fieschi C, Abel L, Puel A, Cobat A, Casanova JL, Zhang Q, Boisson B. J Exp Med. 2021 Nov 1;218(11):e20210566. doi: 10.1084/jem.20210566. Epub 2021 Sep 2. PMID: 34473196; PMCID: PMC8421261.
BTK operates a phospho-tyrosine switch to regulate NLRP3 inflammasome activity. Bittner ZA, Liu X, Mateo Tortola M, Tapia-Abellán A, Shankar S, Andreeva L, Mangan M, Spalinger M, Kalbacher H, Düwell P, Lovotti M, Bosch K, Dickhöfer S, Marcu A, Stevanović S, Herster F, Cardona Gloria Y, Chang TH, Bork F, Greve CL, Löffler MW, Wolz OO, Schilling NA, Kümmerle-Deschner JB, Wagner S, Delor A, Grimbacher B, Hantschel O, Scharl M, Wu H, Latz E, Weber ANR. J Exp Med. 2021 Nov 1;218(11):e20201656. doi: 10.1084/jem.20201656. Epub 2021 Sep 23. PMID: 34554188; PMCID: PMC8480672.
Hematopoietic Stem Cell Transplantation Resolves the Immune Deficit Associated with STAT3-Dominant-Negative Hyper-IgE Syndrome. Harrison SC, Tsilifis C, Slatter MA, Nademi Z, Worth A, Veys P, Ponsford MJ, Jolles S, Al-Herz W, Flood T, Cant AJ, Doffinger R, Barcenas-Morales G, Carpenter B, Hough R, Haraldsson Á, Heimall J, Grimbacher B, Abinun M, Gennery AR. J Clin Immunol. 2021 Jul;41(5):934-943. doi: 10.1007/s10875-021-00971-2. Epub 2021 Feb 1. PMID: 33523338; PMCID: PMC8249289.
Immune checkpoint deficiencies and autoimmune lymphoproliferative syndromes. Gámez-Díaz L, Grimbacher B. Biomed J. 2021 Aug;44(4):400-411. doi: 10.1016/j.bj.2021.04.005. Epub 2021 Apr 19. PMID: 34384744; PMCID: PMC8514790.
Initial presenting manifestations in 16,486 patients with inborn errors of immunity include infections and noninfectious manifestations. Thalhammer J, Kindle G, Nieters A, Rusch S, Seppänen MRJ, Fischer A, Grimbacher B, Edgar D, Buckland M, Mahlaoui N, Ehl S; European Society for Immunodeficiencies Registry Working Party. J Allergy Clin Immunol. 2021 Nov;148(5):1332-1341.e5. doi: 10.1016/j.jaci.2021.04.015. Epub 2021 Apr 23. PMID: 33895260.
The Clinical Utility of Optical Genome Mapping for the Assessment of Genomic Aberrations in Acute Lymphoblastic Leukemia. Lühmann JL, Stelter M, Wolter M, Kater J, Lentes J, Bergmann AK, Schieck M, Göhring G, Möricke A, Cario G, Žaliová M, Schrappe M, Schlegelberger B, Stanulla M, Steinemann D. Cancers (Basel). 2021 Aug 30;13(17):4388. doi: 10.3390/cancers13174388. PMID: 34503197; PMCID: PMC8431583.
The expansion of human T-bethighCD21low B cells is T cell dependent. Keller B, Strohmeier V, Harder I, Unger S, Payne KJ, Andrieux G, Boerries M, Felixberger PT, Landry JJM, Nieters A, Rensing-Ehl A, Salzer U, Frede N, Usadel S, Elling R, Speckmann C, Hainmann I, Ralph E, Gilmour K, Wentink MWJ, van der Burg M, Kuehn HS, Rosenzweig SD, Kölsch U, von Bernuth H, Kaiser-Labusch P, Gothe F, Hambleton S, Vlagea AD, Garcia Garcia A, Alsina L, Markelj G, Avcin T, Vasconcelos J, Guedes M, Ding JY, Ku CL, Shadur B, Avery DT, Venhoff N, Thiel J, Becker H, Erazo-Borrás L, Trujillo-Vargas CM, Franco JL, Fieschi C, Okada S, Gray PE, Uzel G, Casanova JL, Fliegauf M, Grimbacher B, Eibel H, Ehl S, Voll RE, Rizzi M, Stepensky P, Benes V, Ma CS, Bossen C, Tangye SG, Warnatz K. Sci Immunol. 2021 Oct 15;6(64):eabh0891. doi: 10.1126/sciimmunol.abh0891. Epub 2021 Oct 8. PMID: 34623902.
Vulnerability to Meningococcal Disease in Immunodeficiency Due to a Novel Pathogenic Missense Variant in NFKB1. Anim M, Sogkas G, Schmidt G, Dubrowinskaja N, Witte T, Schmidt RE, Atschekzei F. Front Immunol. 2021 Dec 24;12:767188. doi: 10.3389/fimmu.2021.767188. Erratum in: Front Immunol. 2023 May 10;14:1212029. PMID: 35003082; PMCID: PMC8738076.
What can clinical immunology learn from inborn errors of epigenetic regulators? Camacho-Ordonez N, Ballestar E, Timmers HTM, Grimbacher B. J Allergy Clin Immunol. 2021 May;147(5):1602-1618. doi: 10.1016/j.jaci.2021.01.035. Epub 2021 Feb 17. PMID: 33609625.
Publications 2020
Dynamics in protein translation sustaining T cell preparedness. Wolf T, Jin W, Zoppi G, Vogel IA, Akhmedov M, Bleck CKE, Beltraminelli T, Rieckmann JC, Ramirez NJ, Benevento M, Notarbartolo S, Bumann D, Meissner F, Grimbacher B, Mann M, Lanzavecchia A, Sallusto F, Kwee I, Geiger R. Nat Immunol. 2020 Aug;21(8):927-937.
Altered Microbiota, Impaired Quality of Life, Malabsorption, Infection, and Inflammation in CVID Patients With Diarrhoea. van Schewick CM, Nöltner C, Abel S, Burns SO, Workman S, Symes A, Guzman D, Proietti M, Bulashevska A, Moreira F, Soetedjo V, Lowe DM, Grimbacher B. Front Immunol. 2020 Jul 31;11:1654.
Dominant-negative mutations in human IL6ST underlie hyper-IgE syndrome. Béziat V, Tavernier SJ, Chen YH, Ma CS, Materna M, Laurence A, Staal J, Aschenbrenner D, Roels L, Worley L, Claes K, Gartner L, Kohn LA, De Bruyne M, Schmitz-Abe K, Charbonnier LM, Keles S, Nammour J, Vladikine N, Maglorius Renkilaraj MRL, Seeleuthner Y, Migaud M, Rosain J, Jeljeli M, Boisson B, Van Braeckel E, Rosenfeld JA, Dai H, Burrage LC, Murdock DR, Lambrecht BN, Avettand- Fenoel V, Vogel TP; Undiagnosed Diseases Network, Esther CR, Haskologlu S, Dogu F, Ciznar P, Boutboul D, Ouachée-Chardin M, Amourette J, Lebras MN, Gauvain C, Tcherakian C, Ikinciogullari A, Beyaert R, Abel L, Milner JD, Grimbacher B, Couderc LJ, Butte MJ, Freeman AF, Catherinot É, Fieschi C, Chatila TA, Tangye SG, Uhlig HH, Haerynck F, Casanova JL, Puel A. J Exp Med. 2020 Jun 1;217(6):e20191804.“
Characterization of the clinical and immunologic phenotype and management of 157 individuals with 56 distinct heterozygous NFKB1 mutations. Lorenzini T, Fliegauf M, Klammer N, Frede N, Proietti M, Bulashevska A, Camacho-Ordonez N, Varjosalo M, Kinnunen M, de Vries E, van der Meer JWM, Ameratunga R, Roifman CM, Schejter YD, Kobbe R, Hautala T, Atschekzei F, Schmidt RE, Schröder C, Stepensky P, Shadur B, Pedroza LA, van der Flier M, Martínez-Gallo M, Gonzalez-Granado LI, Allende LM, Shcherbina A, Kuzmenko N, Zakharova V, Neves JF, Svec P, Fischer U, Ip W, Bartsch O, Barış S, Klein C, Geha R, Chou J, Alosaimi M, Weintraub L, Boztug K, Hirschmugl T, Dos Santos Vilela MM, Holzinger D, Seidl M, Lougaris V, Plebani A, Alsina L, Piquer-Gibert M, Deyà-Martínez A, Slade CA, Aghamohammadi A, Abolhassani H, Hammarström L, Kuismin O, Helminen M, Allen HL, Thaventhiran JE, Freeman AF, Cook M, Bakhtiar S, Christiansen M, Cunningham-Rundles C, Patel NC, Rae W, Niehues T, Brauer N, Syrjänen J, Seppänen MRJ, Burns SO, Tuijnenburg P, Kuijpers TW; NIHR-BioResource – Rare Diseases Consortium, Warnatz K, Grimbacher B. J Allergy Clin Immunol. 2020 Apr 9:S0091-6749(20)30422-X.
Nonpermissive bone marrow environment impairs early B-cell development in common variable immunodeficiency. Troilo A, Wehr C, Janowska I, Venhoff N, Thiel J, Rawluk J, Frede N, Staniek J, Lorenzetti R, Schleyer MT, Herget GW, Konstantinidis L, Erlacher M, Proietti M, Camacho-Ordonez N, Voll RE, Grimbacher B, Warnatz K, Salzer U, Rizzi M. Blood. 2020 Apr 23;135(17):1452-1457.
Incidence of SCID in Germany from 2014 to 2015 an ESPED* Survey on Behalf of the API*** Erhebungseinheit für Seltene Pädiatrische Erkrankungen in Deutschland (German Paediatric Surveillance Unit) ** Arbeitsgemeinschaft Pädiatrische Immunologie. Shai S, Perez-Becker R, Andres O, Bakhtiar S, Bauman U, von Bernuth H, Classen CF, Dückers G, El-Helou SM, Gangfuß A, Ghosh S, Grimbacher B, Hauck F, Hoenig M, Husain RA, Kindle G, Kipfmueller F, Klemann C, Krüger R, Lainka E, Lehmberg K, Lohrmann F, Morbach H, Naumann-Bartsch N, Oommen PT, Schulz A, Seidemann K, Speckmann C, Sykora KW, von Kries R, Niehues T. J Clin Immunol. 2020 Jul;40(5):708-717. doi: 10.1007/s10875-020-00782-x. Epub 2020 May 26. PMID: 32458183.
Clinical Phenotypes and Immunological Characteristics of 18 Egyptian LRBA Deficiency Patients. Meshaal S, El Hawary R, Adel R, Abd Elaziz D, Erfan A, Lotfy S, Hafez M, Hassan M, Johnson M, Rojas-Restrepo J, Gamez-Diaz L, Grimbacher B, Shoman W, Abdelmeguid Y, Boutros J, Galal N, El-Guindy N, Elmarsafy A. J Clin Immunol. 2020 Jun 6.
A novel NFKBIA variant substituting serine 36 of IκBα causes immunodeficiency with warts, bronchiectasis and juvenile rheumatoid arthritis in the absence of ectodermal dysplasia. Sogkas G, Adriawan IR, Ringshausen FC, Baumann U, Schröder C, Klemann C, von Hardenberg S, Schmidt G, Bernd A, Jablonka A, Ernst D, Schmidt RE, Atschekzei F. Clin Immunol. 2020 Jan;210:108269. 3,969 “doi: 10.1016/j.clim.2019.108269
Dupilumab to treat severe atopic dermatitis in autosomal dominant hyper-IgE syndrome. Sogkas G, Hirsch S, Jablonka A, Witte T, Schmidt RE, Atschekzei F. Clin Immunol 2020;215:108452
Enabling External Inquiries to an Existing Patient Registry by Using the Open Source Registry System for Rare Diseases: Demonstration of the System Using the European Society for Immunodeficiencies Registry.Scheible R, Kadioglu D, Ehl S, Blum M, Boeker M, Folz M, Grimbacher B, Göbel J, Klein C, Nieters A, Rusch S, Kindle G, Storf H. JMIR Med Inform. 2020 Oct 7;8(10):e17420. doi: 10.2196/17420. PMID: 33026355; PMCID: PMC7578818.
Publications 2019
The European Society for Immunodeficiencies (ESID) Registry Working Definitions for the Clinical Diagnosis of Inborn Errors of Immunity. Seidel MG, Kindle G, Gathmann B, Quinti I, Buckland M, van Montfrans J, Scheible R, Rusch S, Gasteiger LM, Grimbacher B, Mahlaoui N, Ehl S; ESID Registry Working Party and collaborators. J Allergy Clin Immunol Pract. 2019 Jul – Aug;7(6):1763-1770. Epub 2019 Feb 15.
Progressive Immunodeficiency with Gradual Depletion of B and CD4⁺ T Cells in Immunodeficiency, Centromeric Instability and Facial Anomalies Syndrome 2 (ICF2). Sogkas G, Dubrowinskaja N, Bergmann AK, Lentes J, Ripperger T, Fedchenko M, Ernst D, Jablonka A, Geffers R, Baumann U, Schmidt RE, Atschekzei F. Diseases. 2019 Apr 4;7(2):34.
Assessing the Functional Relevance of Variants in the IKAROS Family Zinc Finger Protein 1 (IKZF1) in a Cohort of Patients With Primary Immunodeficiency. Eskandarian Z, Fliegauf M, Bulashevska A, Proietti M, Hague R, Smulski CR, Schubert D, Warnatz K, Grimbacher B. Front Immunol. 2019 Apr 16;10:568. eCollection 2019. Erratum in: Front Immunol. 2019 Jun 28;10:1490.
Evaluating laboratory criteria for combined immunodeficiency in adult patients diagnosed with common variable immunodeficiency. von Spee-Mayer C, Koemm V, Wehr C, Goldacker S, Kindle G, Bulashevska A, Proietti M, Grimbacher B, Ehl S, Warnatz K. Clin Immunol. 2019 Jun;203:59-62. Epub 2019 Apr 17.
Corrigendum: Assessing the Functional Relevance of Variants in the IKAROS Family Zinc Finger Protein 1 (IKZF1) in a Cohort of Patients With Primary Immunodeficiency. Eskandarian Z, Fliegauf M, Bulashevska A, Proietti M, Hague R, Smulski CR, Schubert D, Warnatz K, Grimbacher B. Front Immunol. 2019 Jun 28;10:1490. eCollection 2019.
The German National Registry of Primary Immunodeficiencies (2012-2017). El-Helou SM, Biegner AK, Bode S, Ehl SR, Heeg M, Maccari ME, Ritterbusch H, Speckmann C, Rusch S, Scheible R, Warnatz K, Atschekzei F, Beider R, Ernst D, Gerschmann S, Jablonka A, Mielke G, Schmidt RE, Schürmann G, Sogkas G, Baumann UH, Klemann C, Viemann D, von Bernuth H, Krüger R, Hanitsch LG, Scheibenbogen CM, Wittke K, Albert MH, Eichinger A, Hauck F, Klein C, Rack-Hoch A, Sollinger FM, Avila A, Borte M, Borte S, Fasshauer M, Hauenherm A, Kellner N, Müller AH, Ülzen A, Bader P, Bakhtiar S, Lee JY, Heß U, Schubert R, Wölke S, Zielen S, Ghosh S, Laws HJ, Neubert J, Oommen PT, Hönig M, Schulz A, Steinmann S, Schwarz K, Dückers G, Lamers B, Langemeyer V, Niehues T, Shai S, Graf D, Müglich C, Schmalzing MT, Schwaneck EC, Tony HP, Dirks J, Haase G, Liese JG, Morbach H, Foell D, Hellige A, Wittkowski H, Masjosthusmann K, Mohr M, Geberzahn L, Hedrich CM, Müller C, Rösen-Wolff A, Roesler J, Zimmermann A, Behrends U, Rieber N, Schauer U, Handgretinger R, Holzer U, Henes J, Kanz L, Boesecke C, Rockstroh JK, Schwarze-Zander C, Wasmuth JC, Dilloo D, Hülsmann B, Schönberger S, Schreiber S, Zeuner R, Ankermann T, von Bismarck P, Huppertz HI, Kaiser-Labusch P, Greil J, Jakoby D, Kulozik AE, Metzler M, Naumann-Bartsch N, Sobik B, Graf N, Heine S, Kobbe R, Lehmberg K, Müller I, Herrmann F, Horneff G, Klein A, Peitz J, Schmidt N, Bielack S, Groß-Wieltsch U, Classen CF, Klasen J, Deutz P, Kamitz D, Lassay L, Tenbrock K, Wagner N, Bernbeck B, Brummel B, Lara-Villacanas E, Münstermann E, Schneider DT, Tietsch N, Westkemper M, Weiß M, Kramm C, Kühnle I, Kullmann S, Girschick H, Specker C, Vinnemeier-Laubenthal E, Haenicke H, Schulz C, Schweigerer L, Müller TG, Stiefel M, Belohradsky BH, Soetedjo V, Kindle G, Grimbacher B. Front Immunol. 2019 Jul 19;10:1272. eCollection 2019.
A novel NFKBIA variant substituting serine 36 of IkappaBalpha causes immunodeficiency with warts, bronchiectasis and juvenile rheumatoid arthritis in the absence of ectodermal dysplasia. Sogkas G, Adriawan IR, Ringshausen FC, Baumann U, Schroder C, Klemann C, von Hardenberg S, Schmidt G, Bernd A, Jablonka A, Ernst D, Schmidt RE, Atschekzei F. Clin Immunol 2020;210:108269
Late-Onset Antibody Deficiency Due to Monoallelic Alterations in NFKB1. Schröder C, Sogkas G, Fliegauf M, Dörk T, Liu D, Hanitsch LG, Steiner S, Scheibenbogen C, Jacobs R, Grimbacher B, Schmidt RE, Atschekzei F. Front Immunol. 2019 Nov 14;10:2618. eCollection 2019.
Distinct molecular response patterns of activating STAT3 mutations associate with penetrance of lymphoproliferation and autoimmunity. Jägle S, Heeg M, Grün S, Rensing-Ehl A, Maccari ME, Klemann C, Jones N, Lehmberg K, Bettoni C, Warnatz K, Grimbacher B, Biebl A, Schauer U, Hague R, Neth O, Mauracher A, Pachlopnik Schmid J, Fabre A, Kostyuchenko L, Führer M, Lorenz MR, Schwarz K, Rohr J, Ehl S. Clin Immunol. 2019 Nov 23;210:108316.
The architecture of the IgG anti-carbohydrate repertoire in primary antibody deficiencies. Jandus P, Boligan KF, Smith DF, de Graauw E, Grimbacher B, Jandus C, Abdelhafez MM, Despont A, Bovin N, Simon D, Rieben R, Simon HU, Cummings RD, von Gunten S. Blood. 2019 Nov 28;134(22):1941-1950.
Structural Noninfectious Manifestations of the Central Nervous System in Common Variable Immunodeficiency Disorders. van de Ven A, Mader I, Wolff D, Goldacker S, Fuhrer H, Rauer S, Grimbacher B, Warnatz K. J Allergy Clin Immunol Pract. 2019 Dec 16. pii: S2213-2198(19)31026-8.
Long-term outcome of LRBA deficiency in 76 patients after various treatment modalities as evaluated by the immune deficiency and dysregulation activity (IDDA) score. Tesch VK, Abolhassani H, Shadur B, Zobel J, Mareika Y, Sharapova S, Karakoc-Aydiner E, Rivière JG, Garcia-Prat M, Moes N, Haerynck F, Gonzales-Granado LI, Santos Pérez JL, Mukhina A, Shcherbina A, Aghamohammadi A, Hammarström L, Dogu F, Haskologlu S, İkincioğulları AI, Bal SK, Baris S, Kilic SS, Karaca NE, Kutukculer N, Girschick H, Kolios A, Keles S, Uygun V, Stepensky P, Worth A, van Montfrans JM, Peters AM4, Meyts I, Adeli M, Marzollo A, Padem N, Khojah AM, Chavoshzadeh Z, Stefanija MA, Bakhtiar S, Florkin B, Meeths M, Gamez L, Grimbacher B, Seppänen MR, Lankester A, Gennery AR, Seidel MG; Inborn Errors, Clinical, and Registry Working Parties of the European Society for Blood and Marrow Transplantation (EBMT) and the European Society of Immunodeficiencies (ESID). J Allergy Clin Immunol. 2019 Dec 27. pii: S0091-6749(19)32603-X.
Clinical and Immunological Phenotype of Patients With Primary Immunodeficiency Due to Damaging Mutations in NFKB2. Klemann C, Camacho-Ordonez N, Yang L, Eskandarian Z, Rojas-Restrepo JL, Frede N, Bulashevska A, Heeg M, Al-Ddafari MS, Premm J, Seidl M, Ammann S, Sherkat R, Radhakrishnan N, Warnatz K, Unger S, Kobbe R, Hüfner A, Leahy TR, Ip W, Burns SO, Fliegauf M, Grimbacher B. Front Immunol. 2019 Mar 19;10:297. doi: 10.3389/fimmu.2019.00297. PMID: 30941118; PMCID: PMC6435015.