Publications 2023
A TriPPPro-Nucleotide Reporter with Optimized Cell-Permeable Dyes for Metabolic Labeling of Cellular and Viral DNA in Living Cells. Sterrenberg VT, Stalling D, Knaack JIH, Soh TK, Bosse JB, Meier C. Angew Chem Int Ed Engl. 2023 Sep 18;62(38):e202308271.
HSV-1 exploits host heterochromatin for nuclear egress. Lewis HC, Kelnhofer-Millevolte LE, Brinkley MR, Arbach HE, Arnold EA, Sanders S, Bosse JB, Ramachandran S, Avgousti DC. J Cell Biol. 2023 Sep 4;222(9):e202304106.
Spatially resolved protein map of intact human cytomegalovirus virions. Bogdanow B, Gruska I, Mühlberg L, Protze J, Hohensee S, Vetter B, Bosse JB, Lehmann M, Sadeghi M, Wiebusch L, Liu F. Nat Microbiol. 2023 Aug 7.
From the beginnings to multidimensional light and electron microscopy of virus morphogenesis. Saskia Sanders, Yannick Jensen, Rudolph Reimer, Jens B. Bosse, Chapter Two – , Editor(s): Stefan Finke, Dmitry Ushakov, Advances in Virus Research, Academic Press, Volume 116, 2023, Pages 45-88
Viral dew: Phase separation and the formation of viral replication compartments. Bosse JB, Brune W. Viral dew. PLoS Pathog. 2023 Feb 16;19(2):e1011145.
A validated protocol to UV-inactivate SARS-CoV-2 and herpesvirus-infected cells. PLoS One. 2023 May 10;18(5):e0274065. Soh TK, Pfefferle S, Wurr S, von Possel R, Oestereich L, Rieger T, Uetrecht C, Rosenthal M, Bosse JB.
Publications 2022
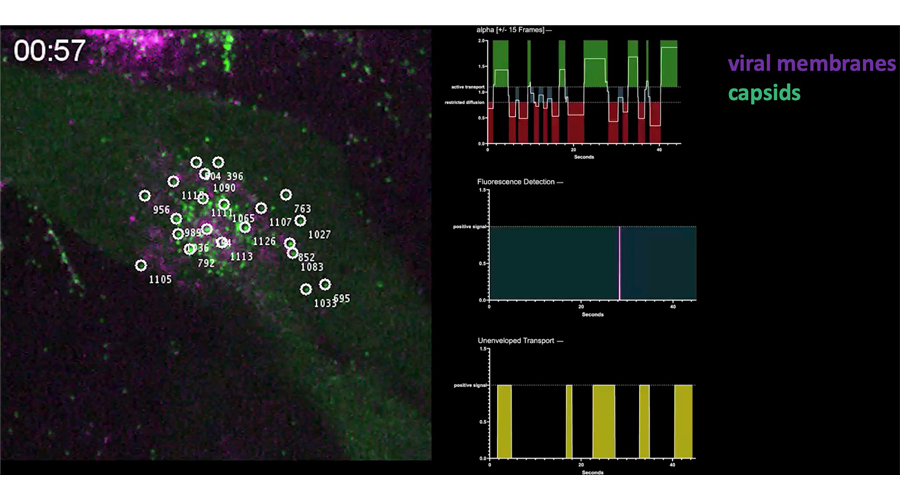
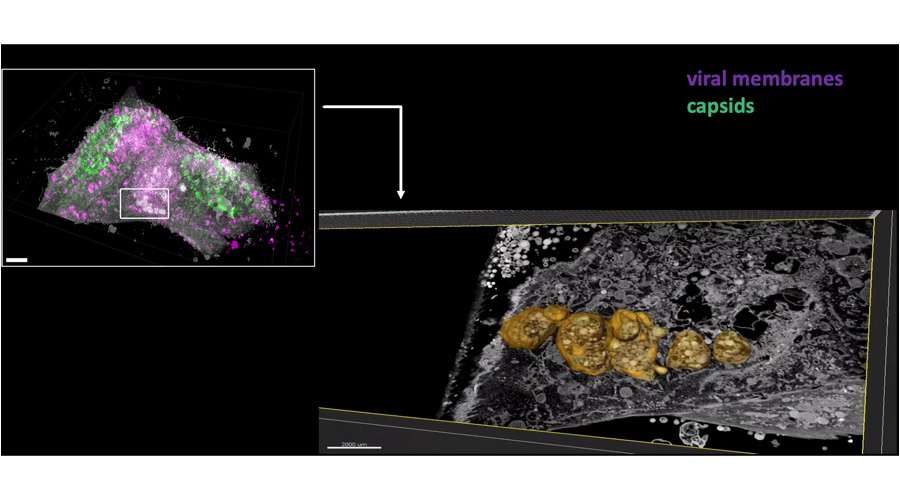
Intermittent bulk release of human cytomegalovirus. Flomm FJ, Soh TK, Schneider C, Wedemann L, Britt HM, Thalassinos K, Pfitzner S, Reimer R, Grünewald K, Bosse JB. PLoS Pathog. 2022 Aug 4
The unconventional way out – Egress of HCMV through multiviral bodies. Wedemann L, Flomm FJ, Bosse JB. Mol Microbiol.
Herpesvirus Replication Compartments: Dynamic Biomolecular Condensates? Enrico Caragliano, Wolfram Brune, Jens B. Bosse, Viruses
Human cytomegalovirus forms phase-separated compartments at viral genomes to facilitate viral replication. Caragliano E, Bonazza S, Frascaroli G, Tang J, Soh TK,Grünewald K, Bosse JB*, Brune W* (*co-corresponding authors, equal contribution) Cell Reports
A Unique Role of the Human Cytomegalovirus Small Capsid Protein in Capsid Assembly. Borst EM, Harmening S, Sanders S, Caragliano E, Wagner K, Lenac Roviš T, Jonjić S, Bosse JB, Messerle M. mBio. 2022 Oct 26;13(5):e0100722. doi: 10.1128/mbio.01007-22. Epub 2022 Sep 6.
Publications 2021
Infection-induced chromatin modifications facilitate translocation of herpes simplex virus capsids to the inner nuclear membrane, V Aho, S Salminen, S Mattola, A Gupta, F Flomm, B Sodeik, JB Bosse, Maija Vihinen-Ranta, PLoS pathogens 17 (12), e1010132
KIR3DS1 directs NK cell-mediated protection against human adenovirus infections. Jung J, Ching W, Baumdick M, Hoffmann-Sieber H, Bosse JB, Koyro TF, Moeller K, Wegner L, Niehrs A, Russu K, Ohms M, Zhang W, Ehrhardt A, Duisters K, Spierings E, Hoelzemer A, Koerner C, Jansen S, Peine S, Koenigs I, Luetgehetmann M, Perez D, Reinshagen K, Lindemanns AA, Altfeld M, Berlderbos ME, Dobner T, Bunders M, (2021), Science Immunology, https://doi.org/10.1126/sciimmunol.abe2942
Concatemeric Broccoli reduces mRNA stability and induces aggregates. Rink MR, Baptista MAP, Flomm FJ, Hennig T, Whisnant AW, Wolf N, Seibel J, Dölken L, Bosse JB, (2021), PLOS One, doi: 10.1371/journal.pone.0244166 Article PLOS ONE 3,2
Identification of African Elephant Polyomavirus in wild elephants and the creation of a vector expressing its viral tumor antigens to transform elephant primary cells. Pearson VR, Bosse JB, Koyuncu OO, Scherer J, Toruno C, Robinson R, et al. (2021) PLoS ONE 16(2): e0244334.
Human Adenovirus Type 5 Infection Leads to Nuclear Envelope Destabilization and Membrane Permeability Independently of Adenovirus Death Protein. Pfitzner S, Bosse JB, Hofmann-Sieber H, Flomm F, Reimer R, Dobner T, Grünewald K, Franken LE, International Journal of Molecular Sciences (IJMS) 2021).
Egress of human cytomegalovirus through multivesicular bodies. Flomm F, Soh TK, Britt H, Schneider C, Reimer R, Thalassinos K, Grünewald K, Bosse JB. bioRxiv 2020
Publications 2020
Fluorescent Protein Tagging of Adenoviral Proeins PV and PIX Reveals ‘Late Virion Accumulation Compartment’. Sieber H, Bosse JB, Franken LE, Grünewald K, Dobner T, (2020), PLOS Pathogens